Research Outline
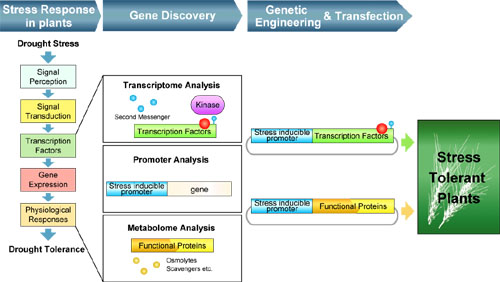
Gene Discovery Research group has been discovering plant genes of which functions are linked to quantitative improvements in plants and those with new functions for minimizing the effects of the environmental stresses to achieve maximum productivity. Many genes with various functions involved in environmental stress responses and tolerance and photosynthesis have been identified for the application to molecular breeding of useful crops with higher yields under global warming. Main targets of the research include not only genes that respond to environmental changes and stimuli and adaptation but also genes involved in photosynthesis and productions of useful metabolites. This group explores key genes involved in improved productivity and abiotic stress tolerance based on integrated genomics, and are analyzing regulatory factors and signaling factors involved in the control of gene expression in response to abiotic environmental stresses. This group also explores regulatory gene networks for the improvement plant productivity. For the research, this group has been constructing a research platform for systematic analysis of plant phenotypes under various growth conditions.
- Discovery of genes, signaling molecules, transporters and metabolites involved in dehydration stress and ABA responses
- Improvement of drought stress tolerance and water use efficiency of crops by international collaboration
- Analysis of chloroplast functions in photosynthesis under stress conditions and discovery of regulatory factors in C4 photosynthesis
- Development of systematic phenotype analysis platform (phenome analysis) for functional analysis of mutated genes
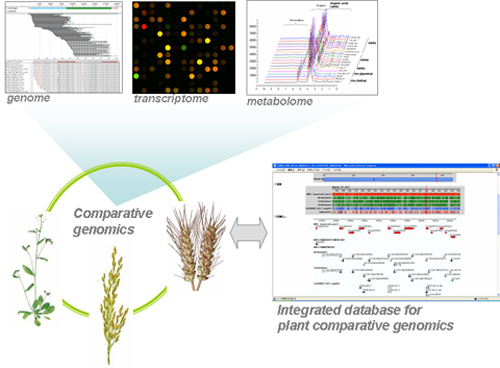
- Comparative genomics and its application to crop improvement
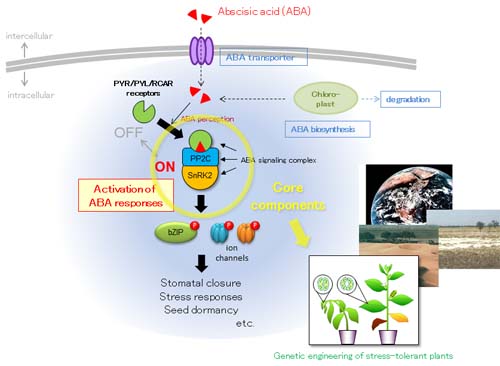
1. Discovery of genes, signaling molecules, transporters and metabolites involved in dehydration stress and ABA responses
To understand regulatory network in environmental stress responses, we focused our work on transcription factors or signal transduction factors involved in plant's responses to drought stress or ABA. We performed functional analyses or biochemical analyses for NAC, bZIP and AP2 transcription factors, or SnRK2 and MAP kinase families.
1-1. CLE25 peptide mutants showed dehydration stress sensitive phenotype.
We found that a peptide moves from roots to leaves, and regulates dehydration stress resistance via abscisic acid biosynthesis.
Takahashi F. et al.,Nature(2018)
Takahashi F. et al.,Nature(2018)

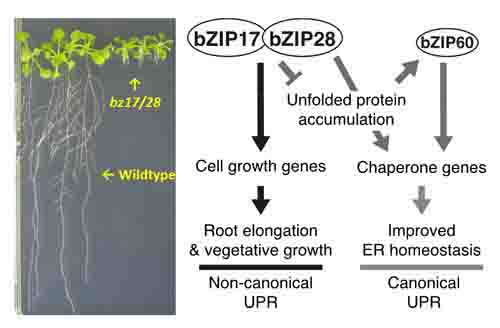
1-2. Defective root growth in the double mutant of bZIP17 and bZIP28 (left). Current working model for extended unfolded protein response regulation via bZIP17 and bZIP28 (upper).
Two ER-anchored transcription factors, bZIP17 and bZIP28 are essential to sustain the normal root elongation in Arabidopsis.
Kim JS. et al., Plant Physiol (2018)
Kim JS. et al., Plant Physiol (2018)
2. Improvement of drought stress tolerance and water use efficiency of crops by international collaboration
To analyze plant responses to abiotic stress, we analyzed functions of stress-inducible genes and metabolites in drought stress responses. Among them, we focused on genes involved in ABA biosynthesis, metabolism and transport.
We found that the intertissue signal transfer of ABA from vascular cells to guard cells.
We found that the intertissue signal transfer of ABA from vascular cells to guard cells.
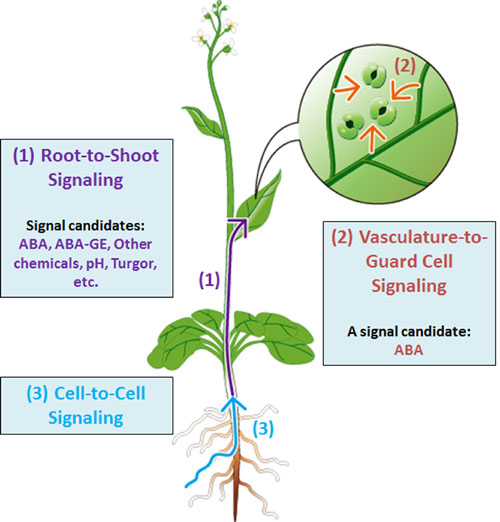
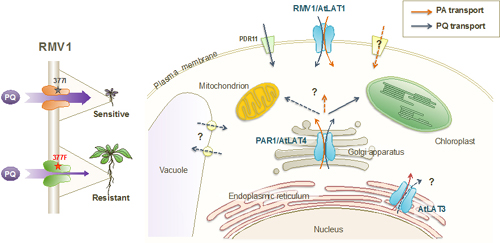
2-1
We developed AtGolS2 overexpressed rice that show resistance to drought stress and increased grain yield in real-world situations.
Selvaraj MG. et al., Plant Biotechnol J (2017)
Selvaraj MG. et al., Plant Biotechnol J (2017)

3. Analysis of chloroplast functions in photosynthesis under stress conditions and discovery of regulatory factors in C4 photosynthesis
We are developing an automatic system for evaluation of plant growth responses to a wide range of environmental conditions.
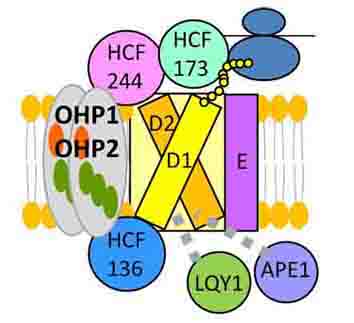
3-1. Schematic model for the role of OHP1-associated proteins during de novo assembly of photosystem II.
We have discovered a new factor involved in the molecular assembly of the photosystem II complex responsible for the photosynthetic reaction.
Myouga F. et al., Plant Physiol (2018)
Myouga F. et al., Plant Physiol (2018)
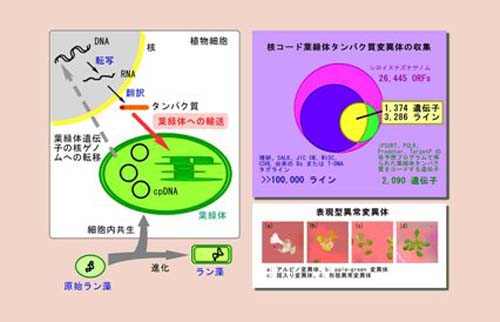
4. Development of systematic phenotype analysis platform (phenome analysis) for functional analysis of mutated genes
To develop environmental stress tolerant crops, we are introducing stress-resistant genes into wheat, rice, and soybean varieties and the field evaluation of stress tolerances in collaboration with international institutes.
5. Comparative genomics and its application to crop improvement
We opened a database on the collection and phenotype analysis of about 2000 Arabidopsis nuclear genes encoding chloroplast proteins for the systematic analysis of chloroplast functions involved in photosynthesis (Chloroplast Function Database II).